Disease worlds models are essential for personalised medicine [Work in progress]
How can world models benefit medicine?
World models might not only be the data engine to power future general AI systems but also a great tool to realise personalised medicine. For example, disease world models should be able to: 1) perform virtual screening to save patients from a preventative perspective; 2) generate virtual personal disease progression trajectories for optimal treatment planning; 3) simulate virtual interventions for helping develop new drugs and planning surgical procedures.
How do we know if we have built a world model for personalised medicine?
A world model should have captured an unparalleled level of detail of the physical world such that the generated artifacts must contain emerging physical properties that are also correct in the physical world and critically significant for clinical purposes. Therefore, toward developing a world model of a disease, the “testing metrics” should be focusing on the artifacts themselves. In the evaluation, we need to look for the emerging properties of the generated medical artifacts. Once the generated medical artifacts satisfy conditions to understand and cure a disease, the system that generates those medical artifacts is a disease world model.
What medical artifacts should we generate?
In history, the emergence of visual intelligence directly led to the Cambrian Explosion. In advancing medicine, medical imaging has also been playing a pivotal role in diagnosis, drug developments and surgery guidance. We should learn from evolution and keep imaging as a centre in the development of world models for diseases. Praticlaly speaking, take the analaogy as the role of videos in the video-based world models for general purposes, that medical imaging can be easily seen as the counterpart in building a disease world model. Especially with the advancement of medical imaging technologies toward micro and smaller scales, different levels of medical scans can help us build a comprehensive disease world model.
Are the current LLM already world models for personalised medicine?
Every day it has become more evident that current LLMs will one day be as good as human doctors at making diagnoses, if enough current clinical representations are embedded and given to the LLMs. However, personalised medicine is far beyond making a diagnosis. Unfortunately, I do not think that the current LLM systems or their future versions are designed for accurate personalised medicine. This is because they can only access sparse and partial clinical representations only in the past, rather than contiously and holistically looking at the whole picture at both the individual and population levels. As a result of the current training pipelines and the current data engineering used to gather the training data, LLMs fundamentally won’t have the ability to generate artifacts that could satisfy the conditions that would make them world models, especially for personalised disease progression and other precision medicine tasks.
World models vs digital twins
The concept of a digital twin was designed to model the physical world in a more abstract and simplified digital version, relying on structural prior knowledge of the physical world. In contrast, a world model does not require prior knowledge; more correct structural representations of the physical world will emerge from the artifacts generated by the world models. In other words, a world model is a more end-to-end approach to digitally reconstruct the physical world. If the recent deep learning revolution has taught us anything, it’s that end-to-end solutions always win in the end.
Contents of this blog:
From the perspective focusing on the emerging properties of the generated medical artifacts, this blog will first lay out the definitions of a Disease World Model, which provides a few future research directions and opportunities. Some early use cases with promising results on lung fibrosis will later be discussed.
Invites for collaborators This is a work in progress, so please feel free to contact me if you find a typo, a mistake, or would like to discuss or collaborate. xumoucheng28@gmail.com.
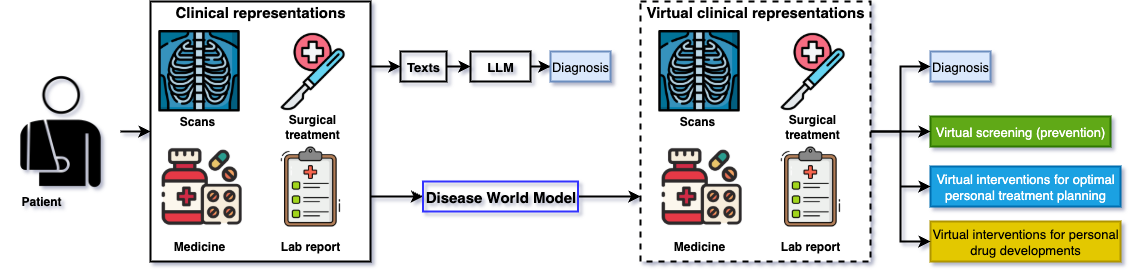
Figure 1: Comparison between Disease World Models and Large Language Models in AI driven healthcare. LLMs can do diagnosis but Disease World Models can do more, even personalised medicine.
Setup for Disease World Model
Before we dive into the formal definitions, we need to define a few terms:
- $X$: synthetic artifacts, as high-dimensional clinical representations (e.g., medical images, medical images + lab reports) of an individual $i \in {1, …, N}$.
- $G$: a generative system that produces a temporal sequence of artifacts ${X_i^t}_{t=0}^{t=E_i}$, where $X_i^t$ is the artifact for individual $i$ at time stamp $t$, $E_i$ is the end point (death time point) of the individual $i$.
- $Y$: real artifacts of an individual $i \in {1, …, N}$ be ${Y_i^t}_{t=S}^{t=E}$, where the real clinical representations are available during the time period between the starting point $t=S$ and the end point $t=E$.
- $M$: a diagnostic agent (e.g., human doctors, clinical LLM) that can map the clinical representations ($X$ and $Y$) to diagnosis. The accuracy of the diagnosis is measured by a metric $\mathcal{L}$.
- $I$: an identification function to recognise the identities of all sequences of the medical artifacts.
- $f_d$: a mapping function that can transform the generated artifact $X$ into another interpretable data format $d$.
- $\mathcal{I}$: a virtual intervention.
Definition 1: Disease World Model
A generative system $G$ is a Disease World Model if its generated artifact sequences ${X_i^t}_{t=0}^{t=E_i}$ satisfy the following properties for all individuals $i$.
- Clinical Comprehensiveness: Each generated artifact $X_i^t$ should contain the complete comprehensive clinical representation of the patient that it can be converted into any data format that is interpretable to human doctors.
- Clinical Reliability: Each generated artifact $X_i^t$ is clinically reliable for all $t$.
- Interventional Validity: Each generated artifact sequence under a virtual intervention is realistic and reliable.
- Individual Characterisability: Each generated artifact sequence ${X_i^t}{t=0}^{t=E_i}$ is _individually characterisable.
Definition 1.1: Clinical Comprehensiveness
An artifact $X_i^t$ is considered clinically comprehensive if it satisfies two conditions:
- Comprehensive Representation: The artifact must encode the complete clinical state of the patient at a given time, rather than a single data modality. Such a comprehensive clinical representation can be seen as the Clinical Platonic Representation of the patient. One such format is medical imaging.
- Functional Convertibility: If the artifact is clinically comprehensive, it must contain the Clinical Platonic Representation of the patient. Therefore, there must exist a set of mapping functions ${f_d}$ capable of converting the artifact $X_i^t$ into any clinically relevant target format $d$ (e.g., medical image, text report, lab reports) that is interpretable by human doctors. Each function performs the transformation $X^t_{i^d} = f_d(X_i^t)$. This ensures that a lot of the existing clinical workflows in the physical world can still be applied on the generated artifacts. It also ensures that the generated artifacts can be directly evaluated for their correctness. For example, if medical images are generated, they should contain the correct key clinical information so that a correct clinical report can be derived from them, and vice versa.
Definition 1.2: Clinical Reliability
An artifact $X_i^t$ is clinically reliable if, for a given diagnostic agent $M$ and accuracy metric $\mathcal{L}$, the following conditions hold:
-
Diagnostic Verifiability: The accuracy of a diagnosis derived from the synthetic artifact should be larger than a threshold $\alpha$:
$L(M(X_i^t)) \ge \alpha, \quad t \in {S, \dots, E}$
In the strictest scenario, the diagnostic accuracy from the synthetic artifact should be at least as good as the real artifacts, more formally, we can define $\alpha \ge L(M(Y_i^t)), \quad t \in {S, \dots, E}$. In less strict scenarios, a threshold $\alpha$ needs to be determined to be clinically accetable. This condition ensures the diagnostic utility of the synthetic artifacts is useable and verifiable.
-
Temporal Consistency: The diagnostic accuracy is a non-decreasing function of time. For any two time points $t_1$ and $t_2$ such that $t_1 < t_2$, we have:
$L(M(X_i^{t_1})) \le L(M(X_i^{t_2})), \quad t_1 < t_2$
Commonly, existing ML models struggle with predicting a time point that is too far away from now on. This condition ensures the diagnostic utility of the synthetic artifacts is reliable, even for future unseen timepoints.
Definition 1.3: Interventional Validity
Let $\mathcal{I}$ be a virtual interaction (e.g., administering a drug, performing surgery, changing of the time) applied at time $t_{\mathcal{I}}$. The generative system $G$ has interventional validity if it can generate a post-intervention sequence that satisfies the following conditions:
- Post-Intervention Plausibility: The generated post-intervention trajectory ${ X_i^t \mid \mathcal{I} }$ is clinically plausible and consistent with established medical knowledge regarding the effects of intervention $\mathcal{I}$. This ensures that the interactions injected from the physical world have meaningful consequences to the artifacts. In the most simple example, there is no artificial intervention, but only one natural intervention, time. In that case, the model should generate artifacts at different arbitrary time points, some of which are verifiable from the clinical records (e.g. past medical scans). In a more comprehensive example, there are artificial interventios such as surgical treatments, the model should generate artifacts at post-treatment stage, some of which are verifiable, such as post-surgery medical scans for checkups.
- Post-Intervention Reliability: Each artifact $X_i^t \mid \mathcal{I}$ generated after the intervention (i.e., for $t \geq t_{\mathcal{I}}$) remains clinically reliable as defined in Definition 1.2. This ensures that the effectiveness of the virtual clinical interactions can be trusted and directly assessed for drug developments and personal treatment planning.
Definition 1.4: Individual Characterisability
A sequence of artifacts ${X_i^t}{t=0}^{t=E_i}$ is _individually characterisable to ensure that the sequence contains a unique signature of the individual’s disease progression, if it satisfies the following conditions:
-
Identifiability: There exists an identification function $I$ that can identify the individual $i$ from their generated sequence with a high probability $\beta$ close to 1.
$\mathbb{P}(I({X_i^t}_{t=0}^{t=E_i}) = i) \ge \beta$
An example of such an ientification function $I$ is an unsupervised contrastive clustering algorithm at a very granular level.
-
Endpoint Fidelity: The generated trajectory for an individual must terminate at a clinically plausible endpoint. When the ground-truth time of death, $E_i^*$, is available for comparison, the sequence’s simulated time of death, $E_i$, must align with it within a predefined, clinically acceptable margin $\delta_E$.
$E_i - E_i^* \le \delta_E$
This ensures the model accurately captures the overall duration and prognostic outcome of the individual’s specific disease progression pattern.
How to train such a Disease World Model
To be updated
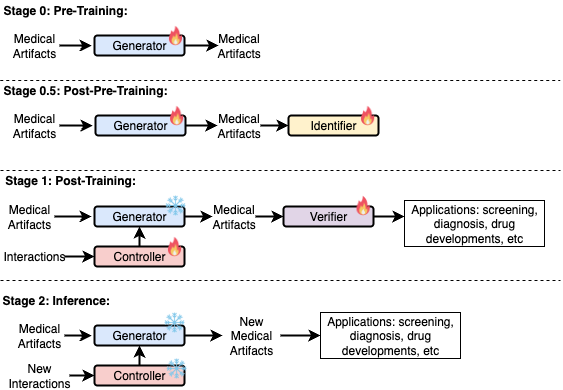
Figure 2: A proposal for the training strategy of a disease world model.
An early attempted use case that is applied to lung fibrosis disease progression modelling
4D-VQGAN: We built an AI model called 4D VQ-GAN for disease progression modelling of the lung fibrosis disease that satisfies the clinical reliability condition of a disease world model. In the context of the disease progression, we can adapt temporal medical imaging as the environments, on the analogy as using the videos as environments in physical world models. As a proof of concept, our early attempt only focus on only one interactive action with the virtual progression trajectories, which is the time. The technical details of this preliminary study can be found in the published conference paper: 4D VQGAN. Given two 3D CT scans of an Idiopathic Pulmonary Fibrosis patient at irregular time points, 4D VQ-GAN can generate synthetic 3D images at any desired time point, effectively modelling a virtual continuous disease progression trajectory for each individual. More importantly, we found that biomarkers derived from the generated CT volumes exhibit a strong clinical correlation with survival outcome, partially satisfying the clinical reliability condition as defined above. This emerging clinical property of the generated CT scans thereby highlights the potential of 4D VQ-GAN for personalized treatment planning and more personalised medicine tasks.
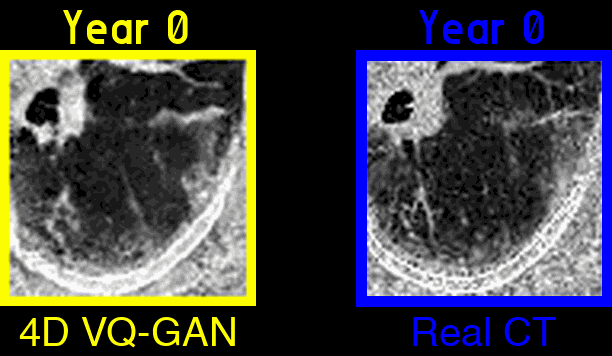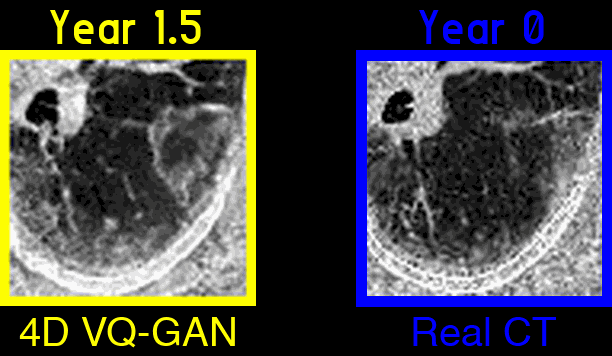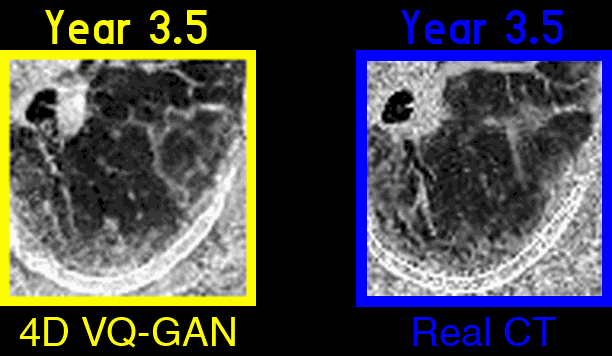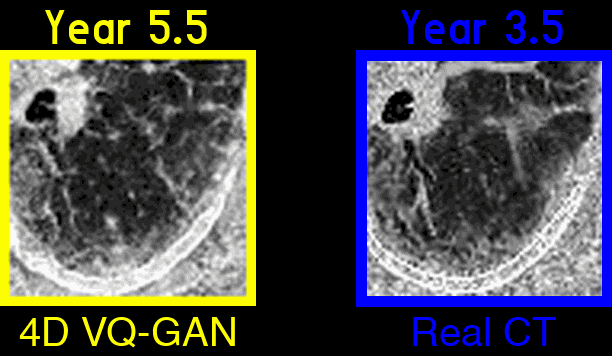
Figure 3: An example of generated synthetic imaging medical artifacts sequence compared against the real sequence. Note that the real scans only contain scans at time point year 0, year2, year 3.5, but the generated scans have more time points at year 0, year 1.5, year 2, year 3.5, year 5.5.
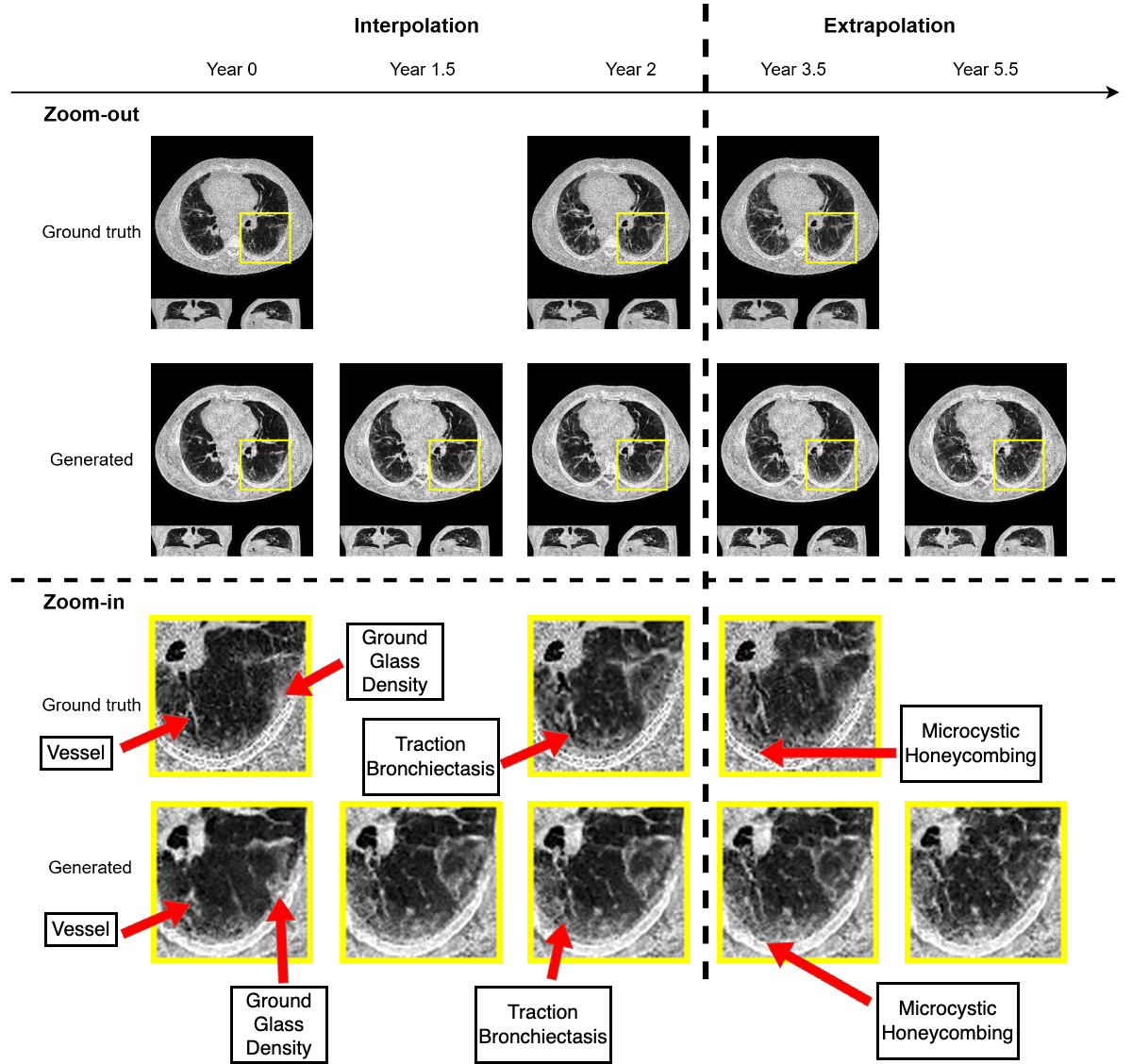
Figure 4: Highlighted key visual features of lung fibrosis in generated imaging artifacts. A zoomed region of the left lower lobe (yellow box) in the real and generated CT scans show comparable amounts of architectural distortion, patterned ground glass opacification and reticulation, all hallmarks of lung fibrosis. The availability of our scans are not uniform across time and across patients, the model is trained on scans at irregular time points
Verifiable Clinical Reliability: We explore our model’s clinical utility using a survival analysis based approach to mimic the clinical workflows. Radiologists track prognostic imaging biomarkers in IPF over time to assess disease progression. Though we lack comprehensive visual scores for all cases, we propose a method that mirrors clinical workflows, including selecting key prognostic biomarkers, analyzing their longitudinal changes, and comparing their prognostic value in synthesized vs. real scans. These extracted imaging biomarkers, along with the covariates, are input into the Cox model to assess their prognostic value in the test dataset. This analysis evaluates the consistency of biomarker trajectories between real and synthetic scans, and explores the potential utility of synthetic scans in tracking changes over time. As shown in the below results, it is very interesting to see that the generated CT images can yield survival outcomes that are sometimes even more accurate than those derived from the real images. However, these results may be overestimated due to the limited sample size.
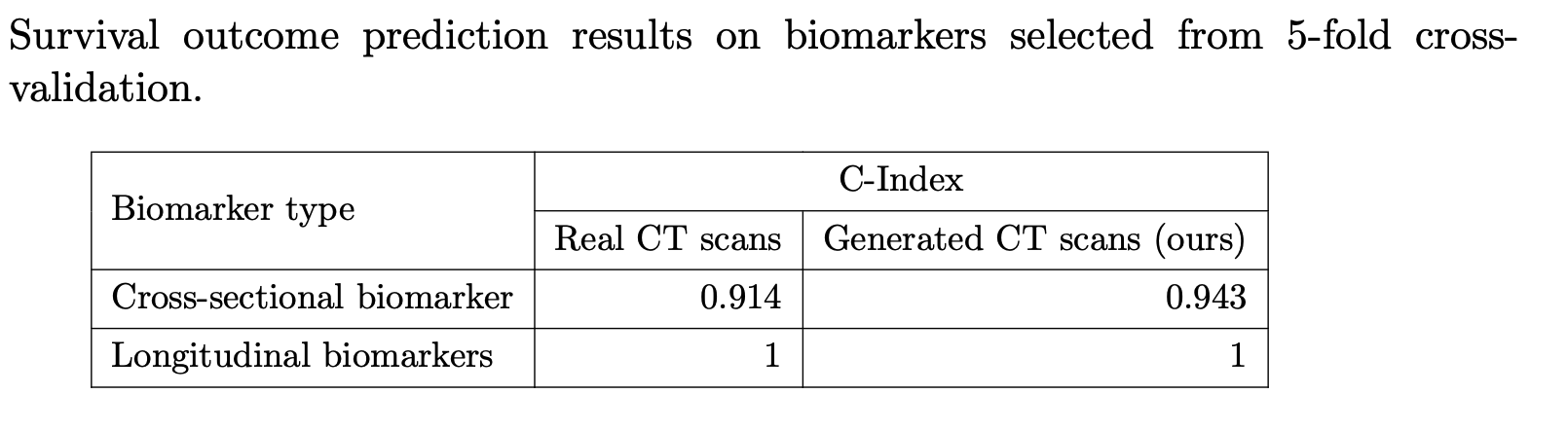
Figure 5: For the cross-sectional imaging biomarker, the C-index using the generated third scans is 0.943. In comparison, using biomarkers derived from real CT scans for survival prediction yields a slightly lower C-index of 0.914. Next, we compute the longitudinal biomarker by evaluating the change in these top five significant biomarkers over the course of one year, both for real and generated CT scans. By inputting these longitudinal biomarkers along with covariates into the Cox model, we obtain C-indices of 1.0 for both real and generated CT scans.
Although the existing 4D VQ-GAN is a step towards the disease world model with promising results, further study is still required to verify if the 4D VQ-GAN also satisfies the clinical reliability condition due to the lack of time and computational resources.
References:
- Genie: Generative Interactive Environments ICML 2024
- Open-Endedness is Essential for Artificial Superhuman Intelligence ICML 2024
- 4D-VQ-GAN: A World Model for Synthesizing Medical Scans at Any Time Point for Personalized Disease Progression Modeling of Idiopathic Pulmonary Fibrosis MIDL 2025
- The Platonic Representation Hypothesis ICML 2024